dersblog
Felzenszwalb Clustering
Using a distance matrix containing pairwise distances (which could use any measure) Felzenszwalb Clustering will print cluster membership per datapoint.
The code is based on Felzenszwalb and Huttenlocher paper and shared C++ code. I simply took out the clustering code from inside the segmentation, so this project does not have any references to image processing, it simply does clustering with any given distance matrix.
http://cs.brown.edu/~pff/segment/
The C++ version reads sparse matrices from mtx (money market
format). Python version can be used as any clustering algorithm found
in scikit-learn, through constructor and fit function.
The main advantage of the algorithm is its ability to use any distance metric, its accuracy (as known from Felzenszwalb image segmentation), its speed, and determining the number of clusters on its own. The algorithm which is at the root of Felzenszwalb clustering is Minimum Spanning Tree which is known to have O(E log E) complexity where E is the number of edges in a graph. In comparison, other clustering methods presented here,
http://scikit-learn.org/stable/modules/clustering.html
such as Affinity Propagation, or Spectral Clustering do not enjoy the same level of performance and cannot handle large datasets.
Building
cd felzclust
make
For Python version simple import is sufficient.
Running
See doc/test.pdf or felzclust/test.py for details. The distance
matrix should be in sparse form, in mtx format. A simple use case
felzclust/felzclust felzclust/simple.mtx 1.0 1
You will see output
point;cluster
0;2
1;2
2;2
3;4
4;4
A bigger example
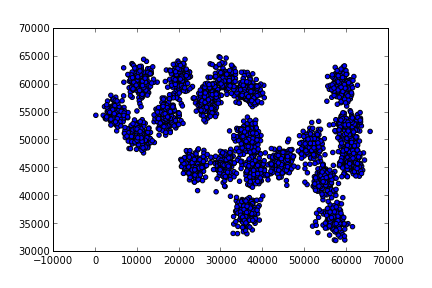
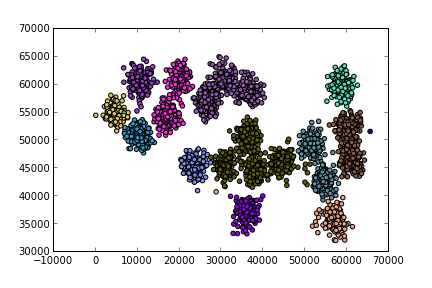
Pure Python
Pure Python version of the code can be found under felz.py and test_felz.py
LICENSE
The software is licensed under GPL v2. See COPYING for details.